
Mycoplasma genitalium Barcelona Team
Institute of Biotechnology and Biomedicine (IBB), Autonomous University of Barcelona
Our research group is located at the Institute of Biotechnology and Biomedicine (IBB), one of the centers of the Research Park at the Universitat Autonoma de Barcelona (UAB). The IBB is a multidisciplinary institute hosting twenty research groups and more than eighty researchers focusing on diverse areas such as Cell and Molecular Biology, Genomics and Proteomics, Immunology, Microbiology and Bioinformatics.
Our team is interested in many aspects of the biology of Mycoplasma genitalium, an emerging sexually transmitted pathogen implicated in urogenital pathologies such as urethritis in men and cervicitis and pelvic inflammatory disease in women. M. genitalium is capable to invade the host target cells by an unknown mechanism, which represents an important asset to avoid the host immune system and establish persistent infections. Numerous studies demonstrate that antibiotic resistance is alarmingly emerging worldwide in this bacterium. Therefore, a better understanding of the virulence mechanisms of this bacterium could facilitate the implementation of more effective antimicrobial approaches. Moreover, M. genitalium possesses one of the smallest genomes known among self-replicating bacteria (580 kb) and constitutes an attractive cell model and a suitable platform for systems biology. So far, we are recognized as a research team leader worldwide in the molecular biology of this pathogen and one of the few groups with the know-how to genetically manipulate this microorganism, with dozens of papers reporting the construction of isogenic mutants by gene replacement aimed to identify and understand the virulence mechanisms and other relevant features of this bacterium.
Adherence, motility and infection
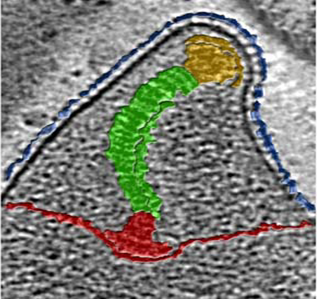
Adhesion of pathogenic bacteria to target cells is a prerequisite for colonization and further infection. M. genitalium possesses a specialized structure known as terminal organelle responsible for adherence to host target cells. The terminal organelle is made by a collection of cytoskeletal proteins that assemble to form a differentiated structure that protrudes from the cell body. The major adhesins of this bacterium, P140 and P110, interact to form a transmembrane complex called NAP, that accumulates at the membrane region that encloses the terminal organelle. In addition, the terminal organelle contains the machinery necessary for gliding motility, which is thought to be important to penetrate the extracellular mucus layer. Our work has been pivotal to understand the specific role of the different components of the terminal organelle in cytadherence and locomotion. In addition, in collaboration with the groups of Ignacio Fita (CSIC, Barcelona) and Achilleas Frangakis (Johann Wolfgang Goethe University, Frankfurt) we are conducting structure-function analyses of several proteins of the terminal organelle, including the two major adhesins, to shed some light on the mechanisms of cell adhesion and motility.
Further readings
García-Morales L, González-González L, Querol E, Piñol J (2016). A minimized motile machinery for Mycoplasma genitalium. Mol Microbiol. 100(1):125-38. (https://doi.org/10.1111/mmi.13305)
Scheffer MP, Gonzalez-Gonzalez L, Seybert A, Ratera M, Kunz M, Valpuesta JM, Fita I, Querol E, Piñol J, Martín-Benito J, Frangakis AS (2017). Structural characterization of the NAP; the major adhesion complex of the human pathogen Mycoplasma genitalium. Mol Microbiol. 105(6):869-879. (https://doi.org/10.1111/mmi.13743)
Burgos R, Pich OQ, Ferrer-Navarro M, Baseman JB, Querol E, Piñol J (2006). Mycoplasma genitalium P140 and P110 cytadhesins are reciprocally stabilized and required for cell adhesion and terminal-organelle development. J Bacteriol. 188(24):8627-37. (https://doi.org/10.1128/JB.00978-06)
Molecular tools
One of the major handicaps in the study of mycoplasmas is the lack of robust genetic tools. For this reason, one of our goals has been the implementation of new technologies for genetic manipulation and genome engineering of M. genitalium. To this end, we developed a minitransposon that has been critical for to perform functional genomics studies and is used in different laboratories world-wide. In addition, we have obtained an OriC based self-replicating plasmid that can be used for heterologous expression of foreign genes in M. genitalium. Remarkably, we have developed a method to obtain unmarked deletions using the Cre-lox technology and, in collaboration with Yo Suzuki (Venter Institute, USA), we have implemented the CRISPR-dCas9 in mycoplasmas.
Further readings
Mariscal AM, Kakizawa S, Hsu JY, Tanaka K, González-González L, Broto A, Querol E, Lluch-Senar M, Piñero-Lambea C, Sun L, Weyman PD, Wise KS, Merryman C, Tse G, Moore AJ, Hutchison CA 3rd, Smith HO, Tomita M, Venter JC, Glass JI, Piñol J, Suzuki Y (2018). Tuning gene activity by inducible and targeted regulation of gene expression in minimal bacterial cells. ACS Synth Biol. 7(6):1538-1552. (https://doi.org/10.1021/acssynbio.8b00028)
Mariscal AM, González-González L, Querol E, Piñol J (2016). All-in-one construct for genome engineering using Cre-lox technology. DNA Res. 23(3):263-70. (https://doi.org/10.1093/dnares/dsw015)
Adaptation and survival within the host
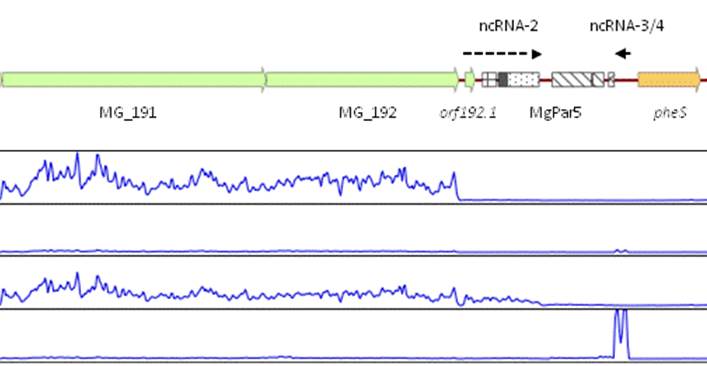
Our lab is also interested in the mechanisms that allow M. genitalum cells to adapt to different environments. These complex adaptative responses are critical to survive within the host and subvert the host immune defenses. To this end, we use genome-wide transcriptional analyses to identify groups of genes that are differentially expressed upon specific conditions. These analyses have revealed the transcriptional networks that operate in M. genitalium and have allowed the identification of novel transcription factors involved in heat stress or regulation of iron homeostasis. In addition, we have identified and characterized an alternative sigma factor (sigma20) controlling homologous recombination in this bacterium. Since recombination represents the main pathway to generate antigenic variation in this bacterium, sigma20 emerges as a novel factor contributing to M. genitalium virulence.
Further readings
Torres-Puig S, Martínez-Torró C, Granero-Moya I, Querol E, Piñol J, Pich OQ (2018). Activation of sigma20-dependent recombination and horizontal gene transfer in Mycoplasma genitalium. DNA Res. Apr 11. (https://doi.org/10.1093/dnares/dsy011)
Torres-Puig S, Broto A, Querol E, Piñol J, Pich OQ (2015). A novel sigma factor reveals a unique regulon controlling cell-specific recombination in Mycoplasma genitalium. Nucleic Acids Res. 43(10):4923-36. (https://doi.org/10.1093/nar/gkv422)
Horizontal gene transfer
In addition, our research studies have demonstrated that M. genitalium cells can transfer DNA horizontally by a novel mechanism that is facilitated by sigma20 overexpression. This DNA transfer system is arguably fundamental for persistence of M. genitalium within the host since it could facilitate a rapid dissemination of successful antigenic variants within the population. Remarkably, this mechanism can be fundamental to explain the rapid spread of antibiotic resistance-mediating mutations. Our laboratory is currently working on the identification of the machinery involved in this unprecedented mechanism of DNA transfer. To this end, our laboratory works in close collaboration with the mycoplasma group in Toulouse to identify the proteins involved in the transfer of DNA and to describe the main features of this unique system.
Further readings
Torres-Puig S, Martínez-Torró C, Granero-Moya I, Querol E, Piñol J, Pich OQ (2018). Activation of sigma20-dependent recombination and horizontal gene transfer in Mycoplasma genitalium. DNA Res. Apr 11. (https://doi.org/10.1093/dnares/dsy011)
Emergence of antibiotic resistance
Azithromycin and moxifloxacin represent the first- and second-line therapies recommended to treat M. genitalium infections. Several studies have shown that resistance to macrolides (azithromycin) and fluoroquinolones (moxifloxacin) is rapidly emerging worldwide. High-throughput sequencing techniques indicate that M. genitalium strains resistant to azithromycin or moxifloxacin present multiple chromosomal SNPs, which precludes the unequivocal identification of key resistance-mediating mutations as well as the assessment of the specific contribution of these SNP to the development of resistance. To gain a full understanding of the SNPs involved in antibiotic resistance in M. genitalium, we are constructing isogenic strains carrying single and multiple mutations in selected genes to test the antibiotic susceptibility of the resulting mutants. This study is carried out in collaboration with Jorgen Jensen (Statens Serum Institut, Copenhagen) and Mateu Espasa (VHIR, Barcelona).
Further readings
Murray GL, Bradshaw CS, Bissessor M, Danielewski J, Garland SM, Jensen JS,Fairley CK, Tabrizi SN (2017). Increasing Macrolide and Fluoroquinolone Resistance in Mycoplasma genitalium. Emerg Infect Dis. 23(5):809-812. (https://doi.org/10.3201/eid2305.161745)
Fernández-Huerta M, Serra-Pladevall J, Barberá MJ, Espasa M (2018). Mycoplasma genitalium and antibiotic resistance in Spain; the need for an effective response against an emerging problem. Enferm Infecc Microbiol Clin. May 28. pii: S0213-005X(18)30183-6. (https://doi.org/10.1016/j.eimc.2018.04.008)
Group Leaders
Jaume Piñol Ribas
Enrique Querol Murillo
Lab members
Josep-Antoni Pérez Pons
Ángel Mozo Villarias
PhD students
Carlos Martínez Torró
Master students
Marina Marcos Silva
Lucía Sánchez Alba
Past members
Luis González González
Raul Burgos Castellanos
Mario Ferrer Navarro
Raquel Planell Cerezo
Contact
jaume.pinyol@uab.cat
Adress
Molecular Biology Laboratory
Institute of Biotechnology and Biomedicine
Campus UAB, Cerdanyola del Valles
08193, Barcelona
Spain
