Do human mycoplasmas rely on iron?
A review article published in the Computational and Structural Biotechnology Journal describes the putative iron-enzymes, transporters, and metalloregulators of four relevant human mycoplasmas and questions the use of iron by these minimal organisms.
Alex Perálvarez-Marín, et al., Computational and Structural Biotechnology Journal, 2021.
Metal utilization in genome-reduced bacteria: do human mycoplasmas rely on iron??
Abstract
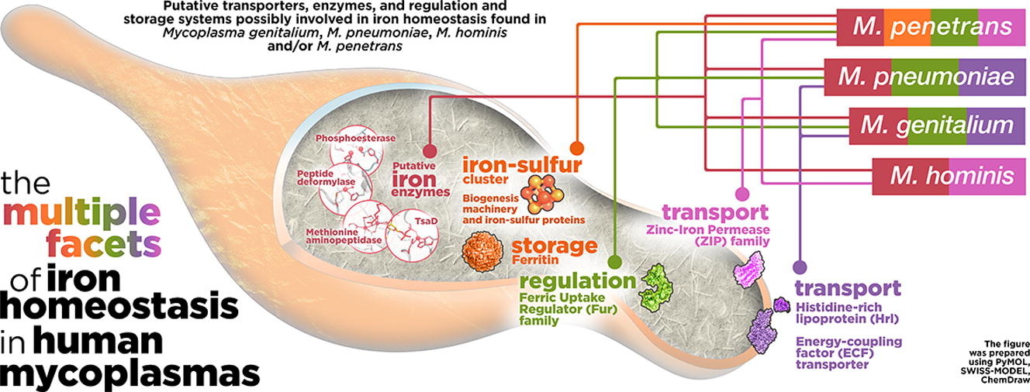
Mycoplasmas are parasitic bacteria with streamlined genomes and complex nutritional requirements. Although iron is vital for almost all organisms, its utilization by mycoplasmas is controversial. Despite its minimalist nature, mycoplasmas can survive and persist within the host, where iron availability is rigorously restricted through nutritional immunity. In this review, we describe the putative iron-enzymes, transporters, and metalloregulators of four relevant human mycoplasmas. This work brings in light critical differences in the mycoplasma-iron interplay. Mycoplasma penetrans, the species with the largest genome (1.36 Mb), shows a more classic repertoire of iron-related proteins, including different enzymes using iron-sulfur clusters as well as iron storage and transport systems. In contrast, the iron requirement is less apparent in the three species with markedly reduced genomes, Mycoplasma genitalium (0.58 Mb), Mycoplasma hominis (0.67 Mb) and Mycoplasma pneumoniae (0.82 Mb), as they exhibit only a few proteins possibly involved in iron homeostasis. The multiple facets of iron metabolism in mycoplasmas illustrate the remarkable evolutive potential of these minimal organisms when facing nutritional immunity and question the dependence of several human-infecting species for iron. Collectively, our data contribute to better understand the unique biology and infective strategies of these successful pathogens.
Full text: https://doi.org/10.1016/j.csbj.2021.10.022


 MDPI
MDPI Alysia Parker
Alysia Parker

 Some of the most important diseases of livestock are caused by mycoplasmas. The article by Nicholas et al., published in Animal Husbandry, Dairy and Veterinary Science provides a comprehensive overview of some of the important issues in animal mycoplasmology:
Some of the most important diseases of livestock are caused by mycoplasmas. The article by Nicholas et al., published in Animal Husbandry, Dairy and Veterinary Science provides a comprehensive overview of some of the important issues in animal mycoplasmology: