
Bordeaux Mollicutes team
UMR 1332 Biologie du Fruit et Pathologie
Our team is involved in fundamental and applied researches studies about plant pathogenic phytoplasmas, animal mycoplasmas and spiroplasmas. The overall objective of the team is to generate fundamental knowledge on the genomes and membranes of mollicutes and the molecular mechanisms that govern the interactions of these “minimal” bacteria with their eukaryotic hosts. This fundamental research is tightly connected with more field-oriented aspects including the integrated epidemiology of grapevine phytoplasmas and mycoplasma genome engineering towards new vaccines, using cutting-edge synthetic biology technologies.

Deciphering and engineering mycoplasma genomes with synthetic biology tools
Using new approaches of synthetic biology, it is now possible to clone a bacterial genome into yeast, to modify it and to transplant it back to a recipient cell. We also adapted several genetic tools for the efficient engineering of mycoplasma genomes cloned in yeast, including a CRISPR/Cas9 system. These approaches are particularly promising for mollicutes for which very few genetic tools are available.
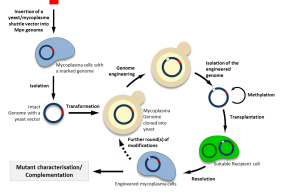
Currently, genome transplantation is still restricted to few species of ruminant pathogenic mycoplasmas belonging or closely related to the so-called Mycoides cluster including Mycoplasma mycoides subsp. capri, Mycoplasma capricolum subsp. capricolum, Mycoplasma leachii. Recently, we succeeded in adapting these methods to the fastest growing mycoplasma, Mycoplasma feriruminatoris. We are involved in several international projects aiming at producing new vaccines against severe animal diseases such as Contagious Bovine Pleuropneumonia and Contagious Caprine Pleuropneumonia (https://www.idrc.ca/en/article/engineering-wild-fast-growing-mycoplasma-bacterium-generate-novel-vaccine-contagious-caprine). Combining comparative genomics and synthetic biology, we also aim at extending genome engineering and transplantation to other bacterial species and use them in applied projects such as construction of new vaccine strains for the control of animal mycoplasma disease and the design of bacterial chassis for biotechnological purposes. This is performed within the frame of several collaborative programs, including the H2020 program MYCOSYNVAC. It is one of our main focus to make genome-scale engineering methods available to many mollicutes and other bacteria.
Recently, our group has discovered a novel mycoplasma virulence factor, based on two surface proteins, termed MiB (Mycoplasma Immunoglobulin Binding) and MiP (Mycoplasma Immunoglobulin Protease). MiB and MiP act in coordination to capture and inactivate the host antibodies by cleaving off their VH domains.
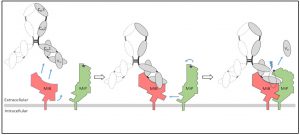
Interestingly, the genes encoding MiB and MiP are part of a large operon containing multiple variants of MiB and MiP, and seven genes encoding an atypical F1-like X0 ATPase whose function is currently unknown. This “MiB-MiP-ATPase” operon is highly conserved throughout the genus Mycoplasma, and is found in almost all the species that infect livestock animals. Given the “antibody killer” role of MiB-MiP, and the widespread distribution of the “MiB-MiP-ATPase” operon, we proposed that this system could be key for the ability of mycoplasmas to evade the host immunity. Taking advantage of our ability to manipulate the genome of Mycoplasma mycoides subsp. capri and using a combination of CryoEM and super-resolution microscopy approaches, we are currently caracterizing this original virulence system of mycoplasmas that might be involved in escaping host immune system. In the long term, we anticipate that this improved fundamental knowledge of mycoplasma virulence will pave the way for the development of new prevention and treatment strategies against mycoplasma diseases in farm animals.
Further readings
Labroussaa F, Lebaudy A, Baby V, Gourgues G, Matteau D, Vashee S, Sirand-Pugnet P, Rodrigue S, Lartigue C. Impact of donor-recipient phylogenetic distance on bacterial genome transplantation. Nucleic Acids Res. 2016 Aug 3. (https://doi.org/10.1093/nar/gkw688)
Arfi Y, Minder L, Di Primo C, Le Roy A, Ebel C, Coquet L, Claverol S, Vashee S, Jores J, Blanchard A, Sirand-Pugnet P. (2016) MIB-MIP is a mycoplasma system that captures and cleaves immunoglobulin G., Proc Natl Acad Sci U S A.,113:5406-11 (https://doi.org/10.1073/pnas.1600546113)
Tsarmpopoulos I, Gourgues G, Blanchard A, Vashee S, Jores J, Lartigue C, Sirand-Pugnet P. In-Yeast Engineering of a Bacterial Genome Using CRISPR/Cas9. ACS Synth Biol. 2016 Jan 15;5(1):104-9. (https://doi.org/10.1021/acssynbio.5b00196)
Grosjean H, Breton M, Sirand-Pugnet P, Tardy F, Thiaucourt F, Citti C, Barré A, Yoshizawa S, Fourmy D, de Crécy-Lagard V, Blanchard A. (2014) Predicting the minimal translation apparatus: lessons from the reductive evolution of mollicutes, PLoS Genet., 10:e1004363. (https://doi.org/10.1371/journal.pgen.1004363)
Lartigue C, Vashee S, Algire MA, Chuang RY, Benders GA, Ma L, Noskov VN, Denisova EA, Gibson DG, Assad-Garcia N, Alperovich N, Thomas DW, Merryman C, Hutchison CA 3rd, Smith HO, Venter JC, Glass JI. Creating bacterial strains from genomes that have been cloned and engineered in yeast. Science. 2009 Sep 25;325(5948):1693-6. (https://doi.org/10.1126/science.1173759)
Integrated epidemiology of phytoplasmoses
Phloem-limited bacterioses, mainly caused by phytoplasmas, are insect-borne incurable diseases. Their control is based on nursery surveillance, elimination of infected plants and insecticide control. Epidemiology and phytoplasma / host relationships constitute our research topics.
We develop molecular diagnostic tools that allow the detection of infected plants for their elimination. We describe new bacterial species, demonstrate their role in the disease, and identify their plant reservoir and insect vectors. Finally, we are using genetic markers to determine the propagation pathways of phytoplasma strains and to predict their epidemic potential, so as to reason insecticide control. Our work focuses on the flavescence dorée and the bois noir phytoplasma diseases of grapevine. We describe the ecological cycles within the vineyards and their environment, and measure the impact of native or introduced insect vectors. The team maintains a collection of phytoplasmas in Madagascar periwinkle in a containment greenhouse.
Our long-term objectives are to identify sources of resistance in grapevine that are useful for varietal improvement, as well as ways to control the spread of flavescence dorée other than insecticides. To bring out these new control strategies, we characterize phytoplasma-host interactions. After phytoplasma ingestion by the insect vector, the steps necessary for transmission include passage through the intestinal barrier, multiplication in the insect tissue and invasion of the salivary glands. For the flavescence dorée phytoplasma we identify its adhesins and their receptors in the insect vector. We are currently inhibiting their expression by RNA interference to confirm the role of these insect proteins in transmission. On the side of the host plant, we characterize the differences in susceptibility to Flavescence dorée within the genus Vitis, in terms of multiplication and diffusion of phytoplasmas in the vine. We inoculate the flavescence dorée phytoplasma with different grape varieties through its leafhopper Scaphoideus titanus. In order to understand the mechanisms involved in this contrasted susceptibility, the variations of the expression of genes involved in defense and metabolism are measured. In addition, upon phytoplasma genome sequencing we have established a list of genes potentially coding for virulence factors, such as surface proteases or proteins secreted by the phytoplasma. We characterize the function of these effectors to discover involvement in the interaction with the host plant or insect vector.
Further readings
Arricau-Bouvery N, Duret S, Dubrana M-P, Batailler B, Desque D, Beven L, Danet J-L, Monticone M, Bosco D, Malembic-Maher S, Foissac X. 2018. Variable Membrane Protein A of Flavescence Doree Phytoplasma Binds the Midgut Perimicrovillar Membrane of Euscelidius variegatus and Promotes Adhesion to Its Epithelial Cells. Applied and Environmental Microbiology 84:e02487-17 (https://doi.org/10.1128/AEM.02487-17)
Eveillard S, Jollard C, Labroussaa F, Khalil D, Perrin M, Desque D, Salar P, Razan F, Hevin C, Bordenave L, Foissac X, Masson JE, Malembic-Maher S. 2016. Contrasting Susceptibilities to Flavescence Doree in Vitis vinifera, Rootstocks and Wild Vitis Species. Frontiers in Plant Sci. 7:1762. (https://doi.org/10.3389/fpls.2016.01762)
Johannesen J, Foissac X, Kehrli P, Maixner M. 2012. Impact of Vector Dispersal and Host-Plant Fidelity on the Dissemination of an Emerging Plant Pathogen. PLoS ONE 7(12): e51809. (https://doi.org/10.1371/journal.pone.0051809)
Fabre A, Danet J-L, Foissac X. 2011. The stolbur phytoplasma antigenic membrane protein gene stamp is submitted to diversifying positive selection. Gene 472:37-41. (https://doi.org/10.1016/j.gene.2010.10.012)
Spiroplasma: a model for membrane structure, function and dynamics studies
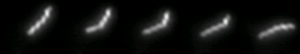
Spiroplasmas are motile, helical mollicutes, primarily infecting arthropods (insects, crustaceans, arachnids…) as final or intermediate (vectors) hosts. Spiroplasmas appear as successful models to understand how these minimal bacteria express the most of their minimal genome at the membrane level. Our main goal is to define the role of membrane organization and structure on pathogenicity, maintenance of cell shape and motility of spiroplasmas. With an interdisciplinary approach combining biophysical, biochemical, cellular and omics studies, several membrane factors involved in spiroplasma pathogenicity could be identified and functionally characterized. Interestingly, our present studies point out the potential role played by post-translational modifications of proteins, and by lipid/protein interactions in spiroplasma pathogenicity. Our research focuses on the etiological agent of Stubborn disease in citrus Spiroplasma citri, which is transmitted in the phloem of the plant by an insect vector, a leafhopper. Therefore, the team also takes advantage of the similarity between the ecological niches of S. citri, which is available in pure cultures, and those of phytoplasmas not yet cultivated: because S. citri can be genetically modified to express on its surface the putative phytoplasma pathogenic factors, this spiroplasma is used successfully in the laboratory as a model for research to decipher the pathogenesis of phytopathogenic mollicutes.
Further readings
Dubrana M-P, Gueguéniat J, Bertin C, Duret S, Arricau-Bouvery N, Claverol S, Lartigue C, Blanchard A, Renaudin J, Béven L (2017) Proteolytic post-translational processing of adhesins in a genome-reduced pathogenic bacterium. J. Mol. Biol. 429 :1889-1902. (https://doi.org/10.1016/j.jmb.2017.05.004)
Renaudin J, Béven L, Batailler B, Desque D, Arricau-Bouvery N, Malembic-Maher S, Foissac X (2015) Heterologous expression and processing of flavescence doree phytoplasma variable membrane protein A in Spiroplasma citri. BMC Microbiol. 15:82. (https://doi.org/10.1186/s12866-015-0417-5)
Duret S, Batailler B, Dubrana M-P, Saillard C, Renaudin J, Béven L, Arricau-Bouvery N (2014) Invasion of insect cells by Spiroplasma citri involves spiralin relocalization and lectin/glycoconjugate-type interactions. Cell Microbiol. 16:1119-1132. (https://doi.org/10.1111/cmi.12265)
Group Leader
Permanent lab members
Brigitte BATAILLER
Jean-Saïd BEY
Martine COINDRE
Delphine DESQUE
Sybille DURET-NURBEL
Christophe GARCION
Denis LACAZE
Thierry LUSSEAU
Isabelle MALDONADO
Sylvie MALEMBIC-MAHER
Annie ROSSARD
Pascal SALAR
PhD students, post docs and contract staff
Laure BATAILLE
Laure DUBOIS
Vincent TALENTON
Contact
pascal.sirand-pugnet@inra.fr
